公司新闻
发布:奥思特科技 发布时间:2019-03-28 来源:本站乙酰化修饰蛋白组学研究利器-乙酰化赖氨酸抗体,Agarose填料
关于 Acetyl Lysine Antibody ,Agarose 的详细介绍
Catalog # Product Description | ICP0388-5mg In order to increase the efficeincy and capacity of the affinity enrichment, higher density of the acetylated lysine antibody is immobilized to beaded agarose. The product could be utilized as an affinity matrix for rapid isolation and purification of the species of proteins or peptide with acetyl lysine residues. |
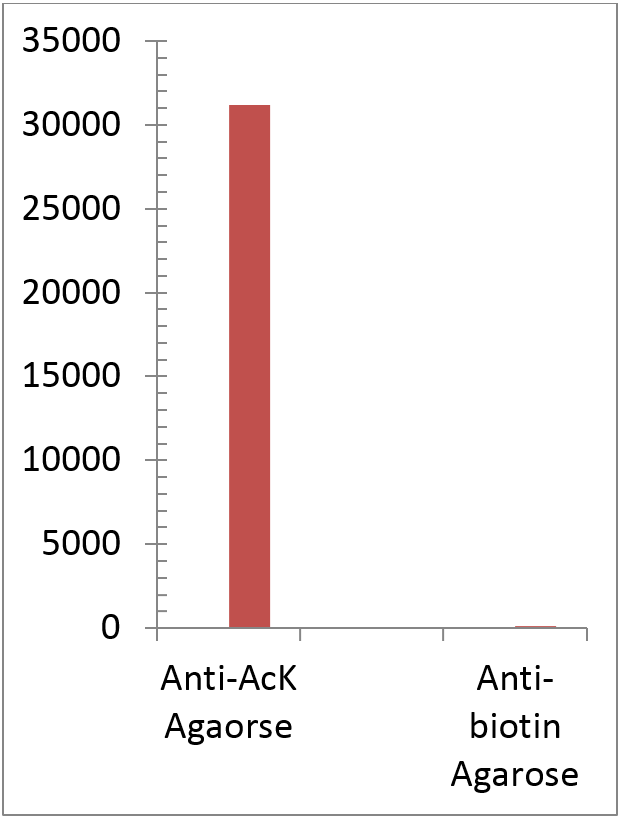
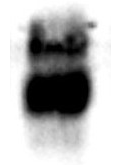
The acetylated protein Signal as determinated by ELISA titration (OD50 titer). Acetlated Proteins were eluted from 1 mL the rabbit anti-acetyllysine Agarose (ICP0388) and immobilized on microrplate followed by detection with mouse anti-Acetylated lysine HRP. Rabbitt anti-Biotin Agarose (1 mL) was utilized as negative control. The maximum binding of acetylated BSA with ICP0388-5MG. -50 µl of ICP0388-5MG were incubated with 1 mg of acetylate BSA in a 1 ml tube for 60 min. - After washing with PBST 4 times, the bound acetylated BSA was eluted with 1 ml of 0.5 M HCl -5 µl / lane of the eluted acetylated BSA was resolved by SDS-PAGE and blot wih monoclonal anti-acetylated lysine (ICP0390) -ECL exposure 3 seconds | |
| Formulation | 0.5 mL beaded agarose suspended in 1 mL slurry | ||
| Antibody Immobilized | 10 mg/mL antibody is covalently linked through amide bonds with NHS activated-SMCC then linked to thiolated agarose beads via thiol ether bonds. | ||
| Specificity | This antibody affinity matrix selectively captures the peptides and proteins with acetylated lysine residues (N-epsilon). There is no cross creation with methylated proteins or mono- and demethylated proteins. | ||
| Binding Capacity | Approximately 0.5-1 mg of acetylated histone/ml. | ||
| Applications | Rapid isolation and purification of peptides or proteins with acetylated lysine residues from cell lysate or protease-digested mixtures. | ||
| Scientific Description | Protein acetylation is a form of post-translational modification known to regulate many diverse biological processes. Detection, isolation and identification of acetylated proteins/peptides is essential in proteomic studies. Affinity chromatography is one of the most efficient and rapid methods to enrich and purify the acetylated species for further MS/MS identification. | ||
| Storage & Stability | Product is stable for 30 days at room temperature. For extended storage, store product at -20°C. Do not aliquot and shake thoroughly before use. | ||
| Product Specific References |
|